Certain RNA molecules in the nerve cells in the brain last a life time without being renewed. Neuroscientists from Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU) have now demonstrated that this is the case together with researchers from Germany, Austria and the USA. RNAs are generally short-lived molecules that are constantly reconstructed to adjust to environmental conditions. With their findings that have now been published in the journal Science, the research group hopes to decipher the complex aging process of the brain and gain a better understanding of related degenerative diseases.
Most cells in the human body are regularly renewed, thereby retaining their vitality. However, there are exceptions: the heart, the pancreas and the brain consist of cells that do not renew throughout the whole lifespan, and yet still have to remain in full working order. “Aging neurons are an important risk factor for neurodegenerative illnesses such as Alzheimer’s,” says Prof. Dr. Tomohisa Toda, Professor of Neural Epigenomics at FAU and at the Max Planck Center for Physics and Medicine in Erlangen. “A basic understanding of the aging process and which key components are involved in maintaining cell function is crucial for effective treatment concepts:”
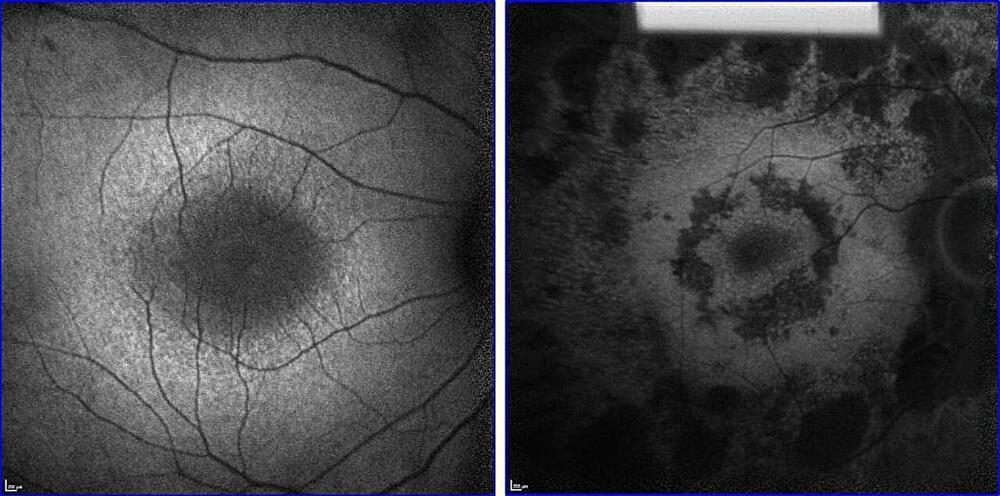
In a joint study conducted together with neuroscientists from Dresden, La Jolla (USA) and Klosterneuburg (Austria), the working group led by Toda has now identified a key component of brain aging: the researchers were able to demonstrate for the first time that certain types of ribonucleic acid (RNA) that protect genetic material exist just as long as the neurons themselves. “This is surprising, as unlike DNA, which as a rule never changes, most RNA molecules are extremely short-lived and are constantly being exchanged,” Toda explains.